arviz_plots.plot_psense_quantities#
- arviz_plots.plot_psense_quantities(dt, alphas=None, quantities=None, mcse=True, var_names=None, filter_vars=None, prior_var_names=None, likelihood_var_names=None, prior_coords=None, likelihood_coords=None, coords=None, sample_dims=None, plot_collection=None, backend=None, labeller=None, aes_by_visuals=None, visuals=None, **pc_kwargs)[source]#
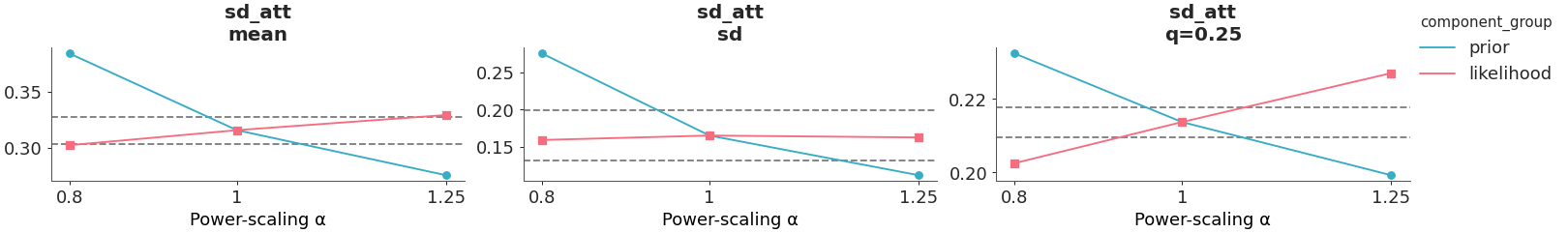
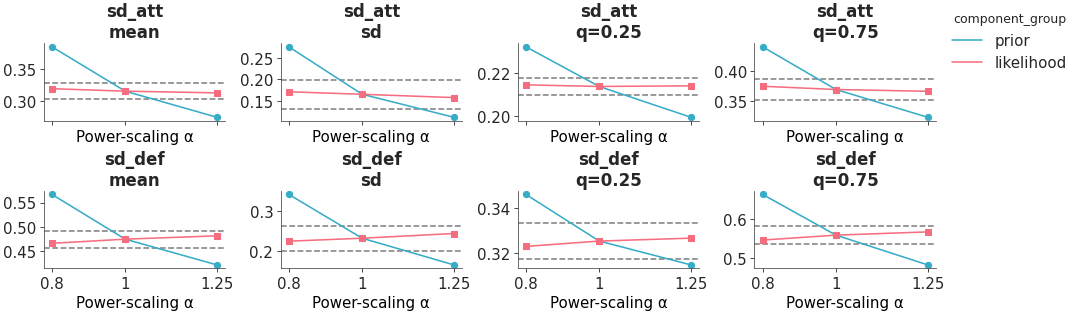
Plot power scaled posterior quantities.
The posterior quantities are computed by power-scaling the prior or likelihood and visualizing the resulting changes, using Pareto-smoothed importance sampling to avoid refitting as explained in [1].
- Parameters:
- dt
xarray.DataTree Input data
- alphas
tupleoffloat Lower and upper alpha values for power scaling. Defaults to (0.8, 1.25).
- quantities
listofstr Quantities to plot. Options are ‘mean’, ‘sd’, ‘median’. For quantiles, use ‘0.25’, ‘0.5’, etc. Defaults to [‘mean’, ‘sd’].
- mcsebool
Whether to plot the Monte Carlo standard error for each quantity. Defaults to True.
- var_names
strorlistofstr, optional One or more variables to be plotted. Prefix the variables by ~ when you want to exclude them from the plot.
- filter_vars{
None, “like”, “regex”}, optional, default=None If None (default), interpret var_names as the real variables names. If “like”, interpret var_names as substrings of the real variables names. If “regex”, interpret var_names as regular expressions on the real variables names.
- prior_var_names
str, optional. Name of the log-prior variables to include in the power scaling sensitivity diagnostic
- likelihood_var_names
str, optional. Name of the log-likelihood variables to include in the power scaling sensitivity diagnostic
- prior_coords
dict, optional. Coordinates defining a subset over the group element for which to compute the log-prior sensitivity diagnostic
- likelihood_coords
dict, optional Coordinates defining a subset over the group element for which to compute the log-likelihood sensitivity diagnostic
- coords
dict, optional - sample_dims
stror sequence of hashable, optional Dimensions to reduce unless mapped to an aesthetic. Defaults to
rcParams["data.sample_dims"]- plot_collection
PlotCollection, optional - backend{“matplotlib”, “bokeh”, “plotly”}, optional
- labeller
labeller, optional - aes_by_visualsmapping of {
strsequence ofstr}, optional Mapping of visuals to aesthetics that should use their mapping in
plot_collectionwhen plotted. Valid keys are the same as forvisuals.- visualsmapping of {
strmapping orFalse}, optional Valid keys are:
prior_markers -> passed to
scatter_xyprior_lines -> passed to
line_xylikelihood_markers -> passed to
scatter_xylikelihood_lines -> passed to
line_xymcse -> passed to
hlineticks -> passed to
set_xtickstitle -> passed to
labelled_titlelegend -> passed to
arviz_plots.PlotCollection.add_legend
- **pc_kwargs
Passed to
arviz_plots.PlotCollection.grid
- dt
- Returns:
References
[1]Kallioinen et al, Detecting and diagnosing prior and likelihood sensitivity with power-scaling, Stat Comput 34, 57 (2024), https://doi.org/10.1007/s11222-023-10366-5
Examples
Select a single parameter, one of the two likelihoods, and plot the mean, standard deviation, and 25th percentile.
>>> from arviz_plots import plot_psense_quantities, style >>> style.use("arviz-variat") >>> from arviz_base import load_arviz_data >>> rugby = load_arviz_data('rugby') >>> plot_psense_quantities(rugby, >>> var_names=["sd_att"], >>> likelihood_var_names=["home_points"], >>> quantities=["mean", "sd", "0.25"])