arviz_plots.plot_ess_evolution#
- arviz_plots.plot_ess_evolution(dt, var_names=None, filter_vars=None, group='posterior', coords=None, sample_dims=None, relative=False, n_points=20, extra_methods=False, min_ess=400, plot_collection=None, backend=None, labeller=None, aes_by_visuals=None, visuals=None, stats=None, **pc_kwargs)[source]#
Plot estimated effective sample size plots for increasing number of iterations.
Roughly speaking, the effective sample size of a quantity of interest captures how many independent draws contain the same amount of information as the dependent sample obtained by the MCMC algorithm. The higher the ESS the better. See [1] for more details.
- Parameters:
- dt
xarray.DataTreeordictof {strxarray.DataTree} Input data. In case of dictionary input, the keys are taken to be model names. In such cases, a dimension “model” is generated and can be used to map to aesthetics.
- var_names
stror sequence ofstr, optional One or more variables to be plotted. Prefix the variables by ~ when you want to exclude them from the plot.
- filter_vars{
None, “like”, “regex”}, defaultNone If None, interpret var_names as the real variables names. If “like”, interpret var_names as substrings of the real variables names. If “regex”, interpret var_names as regular expressions on the real variables names.
- group
str, default “posterior” Group to be plotted.
- coords
dict, optional - sample_dims
stror sequence of hashable, optional Dimensions to reduce unless mapped to an aesthetic. Defaults to
rcParams["data.sample_dims"]- relativebool, default
False Show relative ess in plot
ress = ess / N.- n_points
int, default 20 Number of subsets in the evolution plot.
- extra_methodsbool, default
False Plot mean and sd ESS as horizontal lines.
- min_ess
int, default 400 Minimum number of ESS desired. If
relative=Truethe line is plotted atmin_ess / n_samplesas a curve following themin_ess / ndependency- plot_collection
PlotCollection, optional - backend{“matplotlib”, “bokeh”}, optional
- labeller
labeller, optional - aes_by_visualsmapping of {
strsequence ofstrorFalse}, optional Mapping of visuals to aesthetics that should use their mapping in
plot_collectionwhen plotted. Valid keys are the same as forvisuals.- visualsmapping of {
strmapping orFalse}, optional Valid keys are:
ess_bulk -> passed to
scatter_xyess_bulk_line -> passed to
line_xyess_tail -> passed to
scatter_xyess_tail_line -> passed to
line_xytitle -> passed to
labelled_titlexlabel -> passed to
labelled_xylabel -> passed to
labelled_ymean -> passed to
line_xysd -> passed to
line_xymean_text -> passed to
annotate_xysd_text -> passed to
annotate_xymin_ess -> passed to
line_xy
- statsmapping, optional
Valid keys are:
ess_bulk -> passed to ess, method = ‘bulk’
ess_tail -> passed to ess, method = ‘tail’
mean -> passed to ess, method=’mean’
sd -> passed to ess, method=’sd’
- **pc_kwargs
Passed to
arviz_plots.PlotCollection.wrap
- dt
- Returns:
See also
- Introduction to batteries-included plots
General introduction to batteries-included plotting functions, common use and logic overview
References
[1]Vehtari et al. Rank-normalization, folding, and localization: An improved Rhat for assessing convergence of MCMC. Bayesian Analysis. 16(2) (2021) https://doi.org/10.1214/20-BA1221. arXiv preprint https://arxiv.org/abs/1903.08008
Examples
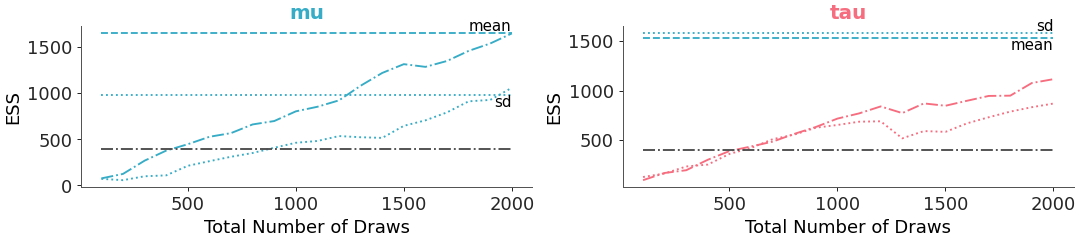
When adding a mapping for color across variables, the same color for a variable gets applied to both the ‘bulk’ and ‘tail’ ess. In such a case, if separate linestyles for ‘bulk’ and ‘tail’ are desired to distinguish them instead of colors (which is what is used by default), then this can be implemented:
>>> from arviz_plots import plot_ess_evolution, style >>> style.use("arviz-variat") >>> from arviz_base import load_arviz_data >>> non_centered = load_arviz_data('non_centered_eight') >>> pc = plot_ess_evolution( >>> non_centered, >>> var_names=["mu", "tau"], >>> extra_methods=True, >>> visuals={ >>> "ess_bulk_line": {"linestyle": "-."}, >>> "ess_tail_line": {"linestyle": ":"}, >>> "ess_bulk": False, >>> "ess_tail": False, >>> }, >>> aes= {"color": ["__variable__"]}, >>> aes_by_visuals={"title": ["color"]}, >>> )
The points and lines for ess ‘bulk’ and ‘tail’ can be individually switched on and off. If only the points are desired, and a situation like the previous example occurs, markers can be used to distinguish between points for ‘bulk’ and ‘tail’:
>>> pc = plot_ess_evolution( >>> non_centered, >>> var_names=["mu", "tau"], >>> extra_methods=True, >>> visuals={ >>> "ess_bulk": {"marker": "x"}, >>> "ess_tail": {"marker": "_"}, >>> }, >>> aes={"color": ["__variable__"]}, >>> aes_by_visuals={"title": ["color"]}, >>> )
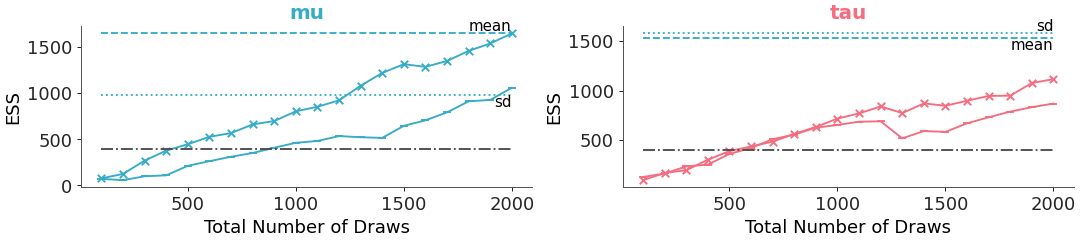
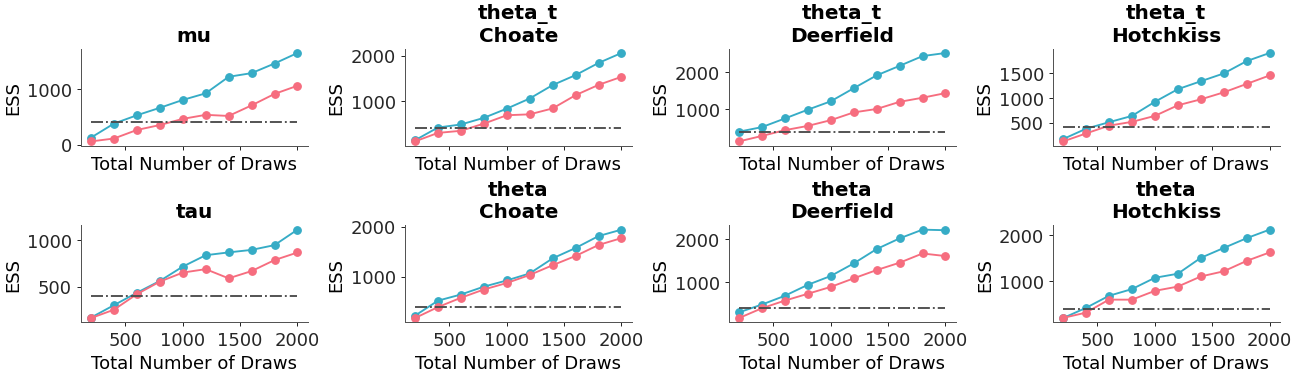
We can add extra methods to plot the mean and standard deviation as lines, and adjust the minimum ess baseline as well:
>>> pc = plot_ess_evolution( >>> non_centered, >>> coords={"school": ["Choate", "Deerfield", "Hotchkiss"]}, >>> extra_methods=True, >>> min_ess=200, >>> )
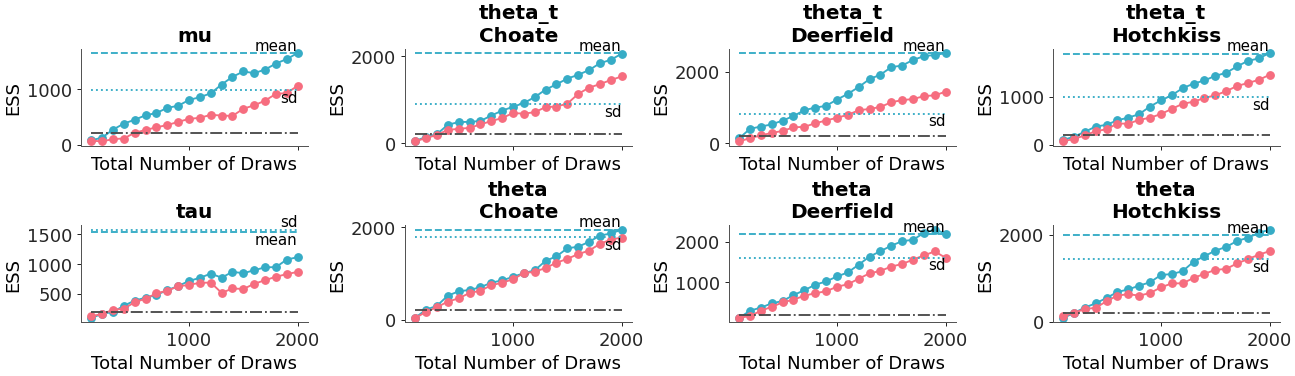
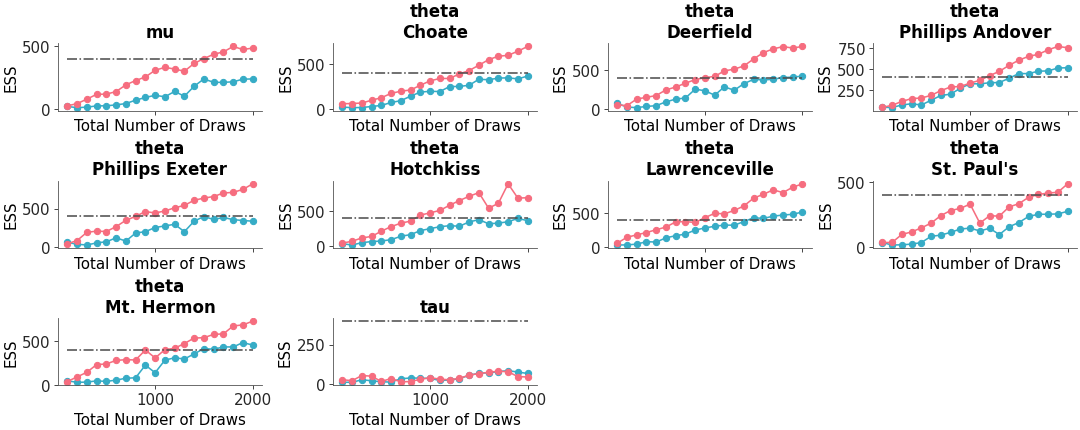
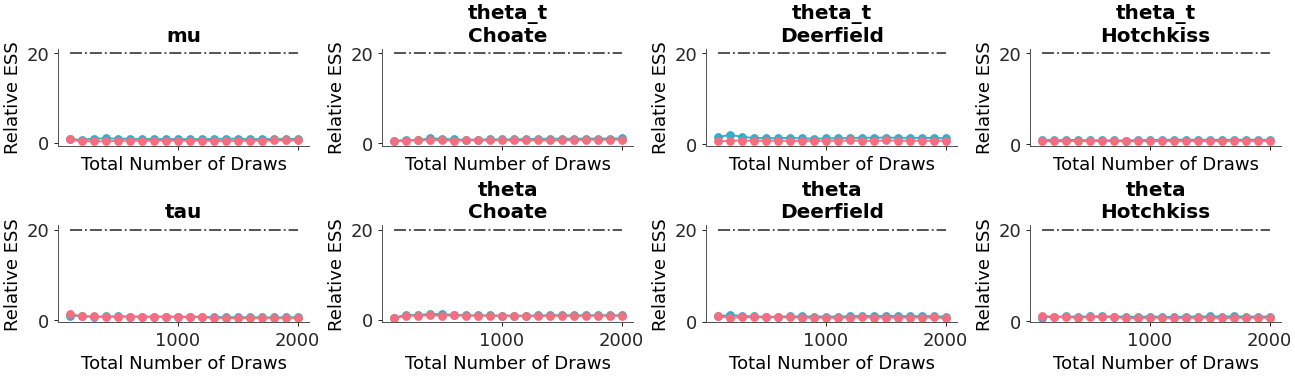
Relative ESS can be plotted instead of absolute:
>>> pc = plot_ess_evolution( >>> non_centered, >>> coords={"school": ["Choate", "Deerfield", "Hotchkiss"]}, >>> relative=True, >>> )
We can also adjust the number of points:
>>> pc = plot_ess_evolution( >>> non_centered, >>> coords={"school": ["Choate", "Deerfield", "Hotchkiss"]}, >>> n_points=10, >>> )